Estimating probabilities
LinXmart uses a probabilistic linkage engine that requires a number of parameters for operation. A set of default parameter values are automatically created when a new Linkage Project is created. To maximise linkage quality, it may be necessary to modify these to best suit the data in question.
LinXmart provides a method to estimate Match () and Non-Match () probabilities, as well as the overall threshold, for a particular linkage. This method uses the Expectation-Maximisation algorithm to estimate these values.
Values are estimated for a particular linkage; that is, for an incoming dataset being both de-duplicated and linked to all other datasets already in the Linkage Project. Both the incoming and pre-existing datasets are included in the analysis to determine the estimated values.
A LinXmart operator carries out this estimation process by loading a Probability Estimation Envelope to the system. The data for linkage must be ingested into the system to carry out this estimation – however it is not linked or maintained within the system. The probability estimation process results in estimated probabilities published and downloadable through the web UI. The LinXmart operator can then update the appropriate match configuration with these probabilities as appropriate, and then load the same data file, this time for linkage.
Ingesting data into LinXmart for estimating probabilities
Probability estimation Envelopes can be created using the LinXmart Simple Envelope Builder. The output of the Build must be set to Probability Estimation. Once created, the Envelope can be loaded into LinXmart using the standard Envelope ingestion method. The system will know how to treat the Envelope based on request type defined in the Envelope's manifest.
Probability Estimation Jobs
After performing the usual validation checks (that the Linkage Project and Event Type exist, and the Event Type is attached to the Linkage Project), the Load Probability Calculation Request job begins.
Load Probability Calculation Request Job
This job parses the data file, and loads valid records into the database.
The parsing that occurs here is the same that occurs with a Load Linkage Request. If the number of records which fail parsing is greater than 5% of the total, the entire datafile is rejected and marked as failed.
Calculate Probabilities Job
This job runs the Expectation-Maximisation (EM) algorithm. This process can be time consuming depending on the size of the data involved. The algorithm itself is iterative; that is, it runs repeatedly, stopping only when it cannot improve the estimates further.
Publish Prob Calcs Envelope
This job publishes the results of the EM process to an Envelope, available to download from the Linkage Project.
Accessing and viewing results from the Probability Calculation process
Once the Publish Prob Calcs Envelope job is completed, the results are downloadable from the web UI. To access these results, go the Project Details page for the Linkage Project in question. On this page, the Probability Estimation pane lists all probability estimation envelopes in the system for that Linkage Project. The estimated correct proportion of matches and threshold score is shown, with further results downloadable by clicking the download icon in the Options column.
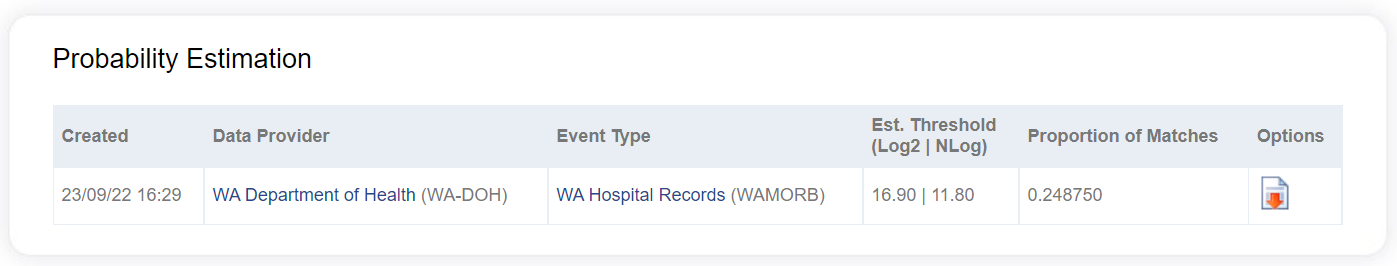
The name of the downloaded zip file contains the code of the Linkage Project, the number of the probability estimation request received by the system, and the datetime stamp of occurrence. E.g. ProbsCalc_APB1_34_20180901030633.zip.
This downloaded zip file contains a number of files that have detailed information regarding the EM process.
| File | Description |
|---|---|
| manifest.xml | The manifest file provides metadata information, outlining that this envelope contains the results of a Probability Estimation request, lists the Linkage Project to which it refers, and lists the other files contained in the envelope. |
| data-summary.txt | This file contains basic summary information on the parameters of the Probability Estimation job, including the number of records and the blocking strategy used. It also contains results of the EM algorithm, including the estimated number and proportion of true matches, and the estimated best threshold choice ("Threshold with highest F-measure (Nlog)"). |
| data-fields.csv | Contains information for each field in linkage, including most importantly, the estimated Match and Non-Match Probabilities for each field. |
| data-fieldcombos.csv | Contains information on the inner workings of the EM algorithm. |
| data-thresholds-nlog.csv | Contain information on the estimated linkage quality found for a range of threshold settings using Natural log for weights. |
| data-thresholds-log2.csv | Contain information on the estimated linkage quality found for a range of threshold settings using Log base 2 for weights. |
The weights found through the Probability Estimation process (located in the data-fields.csv file) for each field can be used by manually entering them through the match configuration process.
Caveats and troubleshooting
Whilst the weight estimation method is a useful technique to estimate and probabilities, it also needs to be used with care.
Firstly, the EM algorithm is not guaranteed to successfully return accurate results in all cases. It works best on administrative datasets with many matches between records. Too few matches and it will fail to produce usable results. The algorithm also cannot tell whether it has succeeded or not. If it has not, then using its estimated weights in linkage will result in poor accuracy.
It is important the Operator recognizes when the returned results look suspect. Generally, probabilities should be in the range of 0.8 to 1.0 for most fields in a dataset. If the results of the EM algorithm suggest probabilities well below that range, then it is likely these results are not useful. There is suggestion in the literature that tighter blocks (that is, changing the blocking values so that less comparisons overall are performed) may resolve this problem.
A second issue is performance. The EM algorithm may perform slowly, more-so as dataset sizes increase. Editing the match configuration for the Linkage Project to change the blocking values so that less comparisons are performed, should reduce the time required for the EM algorithm.